HMDB pathway enrichment
Xiaotao Shen PhD (https://www.shenxt.info/)
Created on 2020-03-28 and updated on 2022-09-19
Source:vignettes/hmdb_pathway_enrichment.Rmd
hmdb_pathway_enrichment.RmdLoad HMDB pathway database
data("hmdb_pathway", package = "metpath")
hmdb_pathway
#> ---------Pathway source&version---------
#> SMPDB & 2021-03-02
#> -----------Pathway information------------
#> 48703 pathways
#> 0 pathways have genes
#> 48642 pathways have proteins
#> 48674 pathways have compounds
#> Pathway class (top 10): Metabolic;primary_pathway
#> get_pathway_class(hmdb_pathway)
#> # A tibble: 12 × 2
#> class n
#> <chr> <int>
#> 1 Disease 20016
#> 2 Disease;primary_pathway 232
#> 3 Drug Action 1
#> 4 Drug Action;primary_pathway 403
#> 5 Drug Metabolism;primary_pathway 64
#> 6 Metabolic 27777
#> 7 Metabolic;primary_pathway 105
#> 8 Physiological 2
#> 9 Physiological;primary_pathway 14
#> 10 Protein;primary_pathway 76
#> 11 Signaling 1
#> 12 Signaling;primary_pathway 12Pathway enrichment
We use the demo compound list from metpath.
data("query_id_hmdb", package = "metpath")
query_id_hmdb
#> [1] "HMDB0000060" "HMDB0000056" "HMDB0000064" "HMDB0000092" "HMDB0000134"
#> [6] "HMDB0000123" "HMDB0000742" "HMDB0000574" "HMDB0000159" "HMDB0000187"
#> [11] "HMDB0000167" "HMDB0000158" "HMDB0000883" "HMDB0000205" "HMDB0000237"
#> [16] "HMDB0000243" "HMDB0000271"Only remain the Metabolic;primary_pathway.
#get the class of pathways
pathway_class =
metpath::pathway_class(hmdb_pathway)
remain_idx = which(unlist(pathway_class) == "Metabolic;primary_pathway")
remain_idx
#> SMP0000055 SMP0000067 SMP0000072 SMP0000015 SMP0000057 SMP0000018 SMP0000029
#> 1 2 3 4 5 6 7
#> SMP0000045 SMP0000009 SMP0000020 SMP0000007 SMP0000123 SMP0000066 SMP0000073
#> 8 9 10 11 12 13 14
#> SMP0000028 SMP0000465 SMP0000012 SMP0000013 SMP0000036 SMP0000468 SMP0000449
#> 15 16 17 18 19 20 21
#> SMP0000054 SMP0000051 SMP0000053 SMP0000064 SMP0000124 SMP0000455 SMP0000071
#> 22 23 24 25 26 27 28
#> SMP0000037 SMP0000129 SMP0000010 SMP0000027 SMP0000126 SMP0000459 SMP0000070
#> 29 30 31 32 33 34 35
#> SMP0000076 SMP0000445 SMP0000021 SMP0000065 SMP0000041 SMP0000450 SMP0000008
#> 36 37 38 39 40 41 42
#> SMP0000044 SMP0000075 SMP0000068 SMP0000127 SMP0000464 SMP0000025 SMP0000444
#> 43 44 45 46 47 48 49
#> SMP0000023 SMP0000032 SMP0000050 SMP0000017 SMP0000060 SMP0000031 SMP0000033
#> 50 51 52 53 54 55 56
#> SMP0000005 SMP0000130 SMP0000006 SMP0000011 SMP0000039 SMP0000035 SMP0000040
#> 140 141 142 143 144 145 146
#> SMP0000034 SMP0000016 SMP0000058 SMP0000048 SMP0000128 SMP0000457 SMP0000466
#> 148 149 150 151 152 153 154
#> SMP0000030 SMP0000462 SMP0000004 SMP0000024 SMP0000043 SMP0000046 SMP0000052
#> 155 156 157 158 159 160 161
#> SMP0000059 SMP0000063 SMP0000074 SMP0000355 SMP0000452 SMP0000456 SMP0000463
#> 162 163 164 165 166 167 168
#> SMP0000467 SMP0000479 SMP0000480 SMP0000481 SMP0000482 SMP0000477 SMP0000478
#> 169 170 171 172 173 453 454
#> SMP0000654 SMP0000715 SMP0000716 SMP0014212 SMP0015896 SMP0020986 SMP0029731
#> 623 684 685 725 2406 7488 16221
#> SMP0030406 SMP0030880 SMP0121055 SMP0121057 SMP0121060 SMP0121123 SMP0121131
#> 16889 16890 48697 48698 48699 48700 48703
hmdb_pathway =
hmdb_pathway[remain_idx]
hmdb_pathway
#> ---------Pathway source&version---------
#> SMPDB & 2021-03-02
#> -----------Pathway information------------
#> 105 pathways
#> 0 pathways have genes
#> 99 pathways have proteins
#> 98 pathways have compounds
#> Pathway class (top 10): Metabolic;primary_pathway
#> result =
enrich_hmdb(query_id = query_id_hmdb,
query_type = "compound",
id_type = "HMDB",
pathway_database = hmdb_pathway,
only_primary_pathway = TRUE,
p_cutoff = 0.05,
p_adjust_method = "BH",
threads = 3)Check the result:
result
#> ---------Pathway database&version---------
#> SMPDB & 2021-03-02
#> -----------Enrichment result------------
#> 98 pathways are enriched
#> 8 pathways p-values < 0.05
#> Glycine and Serine Metabolism;Methionine Metabolism;Phenylalanine and Tyrosine Metabolism;Homocysteine Degradation;Ammonia Recycling ... (only top 5 shows)
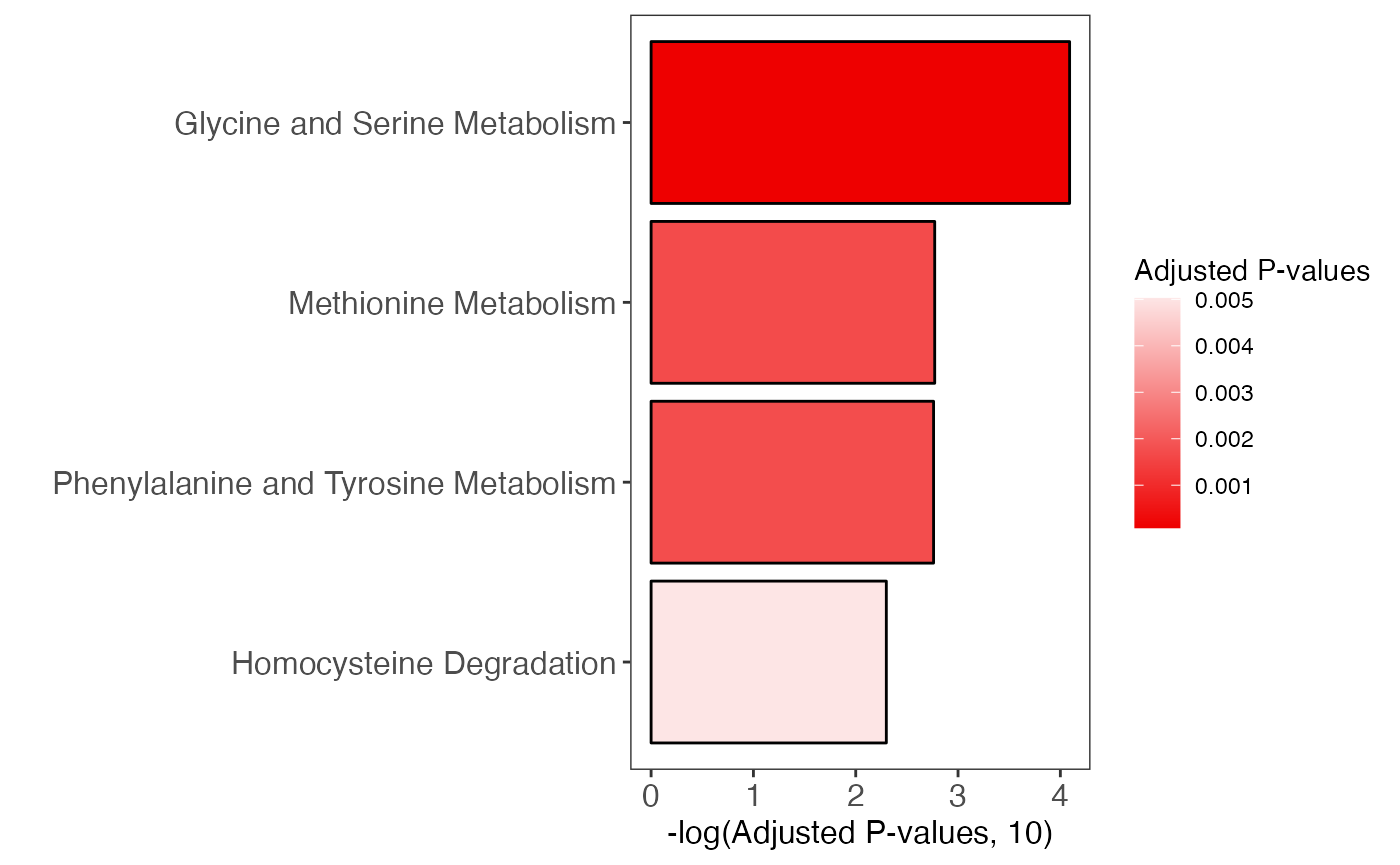
#> Plot to show pathway enrichment
enrich_bar_plot(
object = result,
x_axis = "p_value_adjust",
cutoff = 0.05,
top = 10
)
enrich_scatter_plot(object = result)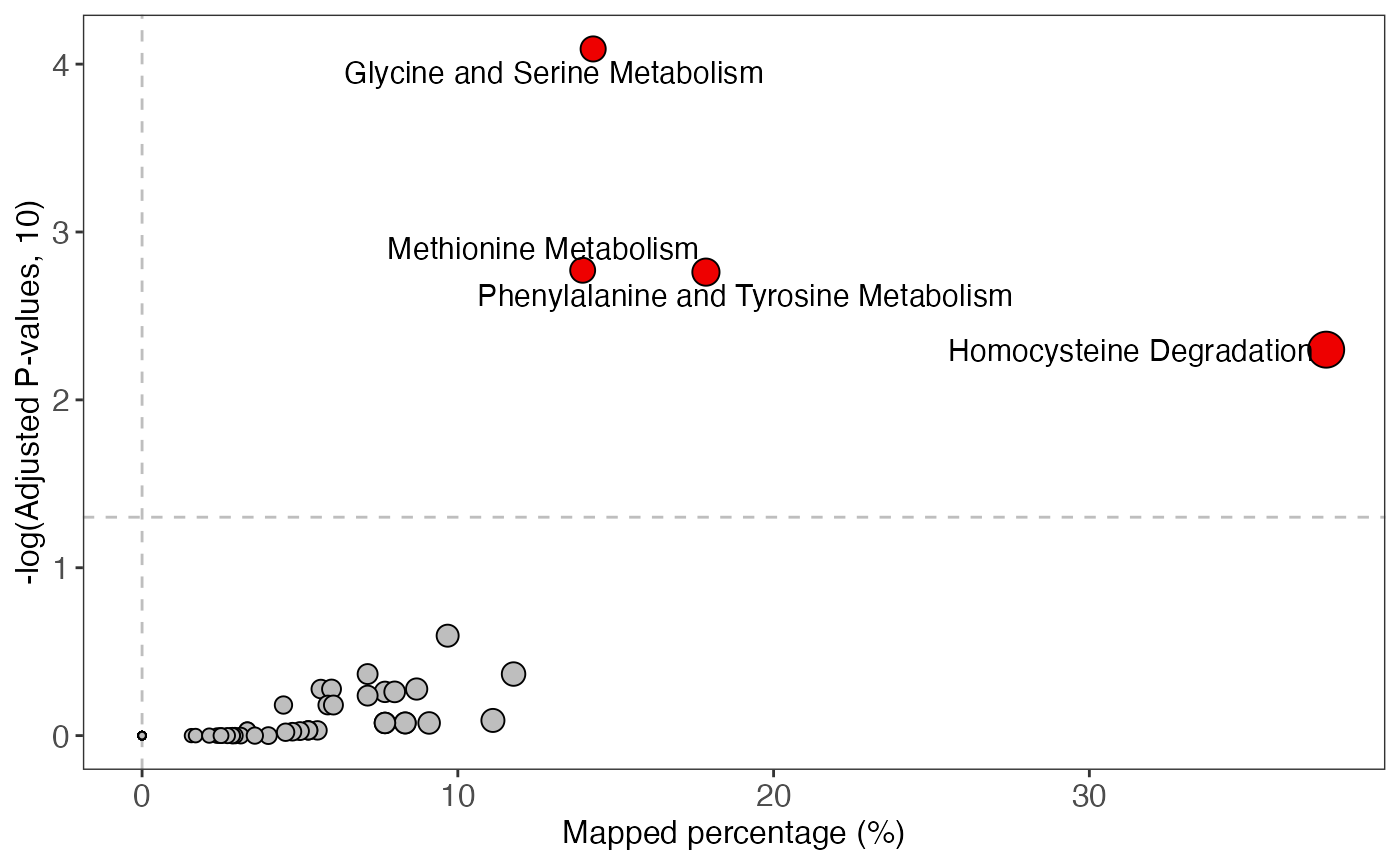
enrich_network(object = result)